
Calculates the ratio of the average pairwise distance among genes within the same gene set and genes within different gene sets.
| Parameters: |
|
|---|---|
| Type : | collection: list of lists |
| Returns: | \(\rho(\mathbf{P})\) (float) - see definition below. |
Then the average pairwise distance ratio \(\rho(\mathbf{P}) = \alpha'(\mathbf{P})-\beta'(\mathbf{P})\). Lower values of \(\rho(\mathbf{P})\) indicate stronger collections.
Note: if no path exists between any genes in the collection, the average pairwise distance ratio will go to infinity since the distance between two nodes in different components (or nodes not in the graph) is ill-defined.
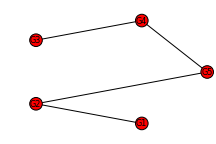
>>> import networkx as nx
>>> collection = [["G1", "G2"], ["G3", "G4"]]
>>> G = nx.Graph()
>>> G.add_edges_from([("G1", "G2"), ("G3", "G4"), ("G2", "G5"), ("G4", "G5")])
>>> nx.draw_circular(G, node_size=125, font_size=8)
>>> dist_within = sum_dist(G, pairs_within(collection))
>>> alpha_prime = float(dist_within) / float(len(pairs_within(collection)))
>>> (dist_within, alpha_prime)
(2, 1.0)
>>> dist_between = sum_dist(G, pairs_between(collection))
>>> beta_prime = float(dist_between) / float(len(pairs_between(collection)))
>>> (dist_between, beta_prime)
(12, 3.0)
>>> avg_pair_dist_ratio(G, collection)
0.33
See also: direct_interactions_test(), eval_gene_sets_by_interactions().