[1]:
# This cell is to allow automatic notebook generation for docs
# You may want to comment this out if you have paste3 installed
import sys
from pathlib import Path
sys.path.insert(0, str(Path.cwd().parent.parent.parent / "src"))
Using the PASTE/PASTE2 algorithm through a unifying API
This noteook highlights the creation of slices (the Slice class) and datasets (the AlignmentDataset class), and the usage of the pairwise_align and center_align methods of the AlignmentDataset class.
This notebook primarily highlights how you would use the ``paste3`` package in either the ``PASTE`` (i.e. full alignment) mode, or the ``PASTE2`` (i.e. partial alignment) mode.. This API also closely reflects how our napari plugin works under the hood, so getting familiar with this API will also help you get familiar with the various options available to you in the plugin.
The paste3.dataset module provides an easy-to-use API to access input datasets to the paste3 alignment algorithms.
The Slice class is a thin layer on top of an AnnData class, and an AlignmentDataset class is a collection of Slice objects.
[2]:
from paste3.dataset import AlignmentDataset
from paste3.napari.data.ondemand import get_file
Individual Slice objects are created by providing a path to an .h5ad file. Each .h5ad file is expected to contain an AnnData object, and is internally read using a scanpy.read_h5ad.
Here we download and cache a few .h5ad files locally using a paste3.napari.data.ondemand.get_file call. These are the files available as the Sample Data in the paste3 napari plugin.
[3]:
slice_files = [Path(f) for f in get_file("paste3_sample_patient_2_")]
Downloading data from 'https://dl.dropboxusercontent.com/scl/fi/zq0dlcgjaxfe9fqbp0hf4/paste3_sample_patient_2_slice_0.h5ad?rlkey=sxj5c843b38vd3iv2n74824hu&st=wcy6oxbt&dl=1' to file '/home/runner/.cache/paste3/paste3_sample_patient_2_slice_0.h5ad'.
Downloading data from 'https://dl.dropboxusercontent.com/scl/fi/a5ufhjylxfnvcn5sw4yp0/paste3_sample_patient_2_slice_1.h5ad?rlkey=p6dp78qhz6qrh0ut49s7b3fvj&st=tyamjq8b&dl=1' to file '/home/runner/.cache/paste3/paste3_sample_patient_2_slice_1.h5ad'.
Downloading data from 'https://dl.dropboxusercontent.com/scl/fi/u7aaq9az8sia26cn4ac4s/paste3_sample_patient_2_slice_2.h5ad?rlkey=3ynobd5ajhlvc7lwdbyg0akj1&st=0l2nw8i2&dl=1' to file '/home/runner/.cache/paste3/paste3_sample_patient_2_slice_2.h5ad'.
A dataset is created using the paths to the individual slices.
[4]:
dataset = AlignmentDataset(file_paths=slice_files)
Any individual slice can be rendered in a jupyter notebook by simply typing the slice variable name in a cell, which renders the slice using the scanpy library. (Note: This is roughly equivalent to doing scanpy.pl.spatial(<slice>.adata, ..))
[5]:
dataset.slices[0]
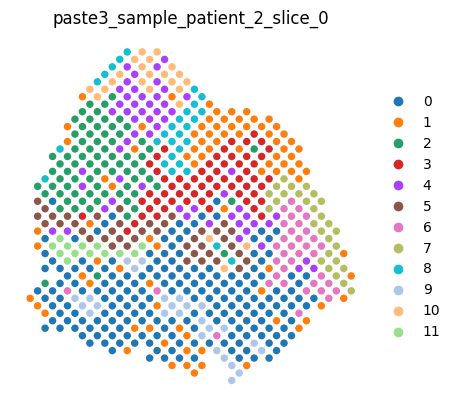
[5]:
<paste3.dataset.Slice at 0x7facc8874770>
An entire dataset can be rendered by typing the dataset variable name in a cell, which renders each slice in order.
[6]:
dataset
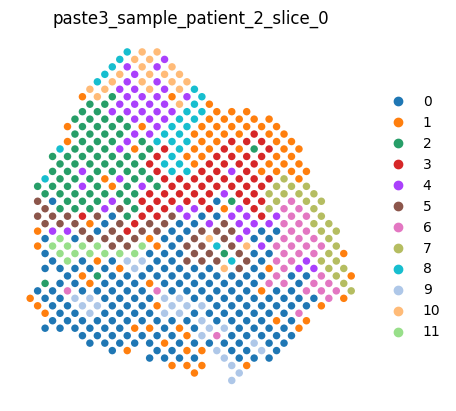
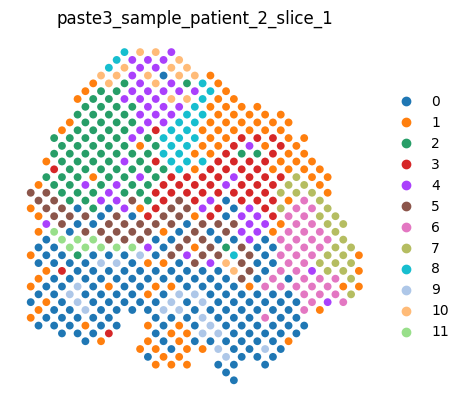
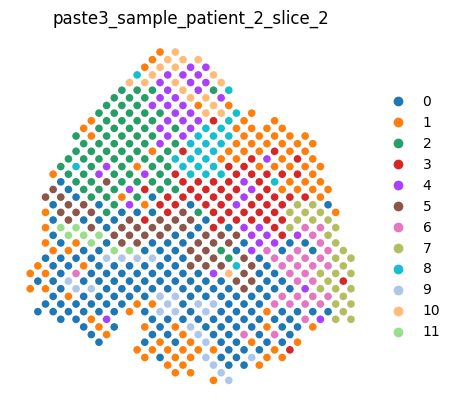
[6]:
<paste3.dataset.AlignmentDataset at 0x7facc8875070>
Center Aligning a Dataset
A dataset object can be center aligned in 2 steps:
Find the "center slice" (or the "consensus slice") and similarity matrix between spots using the
<dataset>.find_center_slicemethod. This is a time consuming step and benefits from being run on a GPU-enabled environment.Use these values to center align the dataset using the
<dataset>.center_alignmethod.
The first returned value is the aligned dataset, along with other useful information (rotations/translations). Here we ignore all returned values except the first one.
Center alignment is explained in detail in the Paste paper.
[7]:
center_slice, pis = dataset.find_center_slice()
aligned_dataset, *_ = dataset.center_align(center_slice=center_slice, pis=pis)
(INFO) (dataset.py) (10-Jan-25 17:13:59) Finding center slice
(INFO) (paste.py) (10-Jan-25 17:13:59) GPU is not available, resorting to torch CPU.
(INFO) (paste.py) (10-Jan-25 17:13:59) Solving Center Mapping NMF Problem.
(INFO) (paste.py) (10-Jan-25 17:14:27) Iteration: 0
(INFO) (paste.py) (10-Jan-25 17:14:27) Solving Pairwise Slice Alignment Problem.
(INFO) (paste.py) (10-Jan-25 17:14:27) Slice 0
(INFO) (paste.py) (10-Jan-25 17:14:27) Slice 1
(INFO) (paste.py) (10-Jan-25 17:14:28) Slice 2
(INFO) (paste.py) (10-Jan-25 17:14:29) center_ot done
(INFO) (paste.py) (10-Jan-25 17:14:29) Solving Center Mapping NMF Problem.
(INFO) (paste.py) (10-Jan-25 17:14:38) Objective -142.8180211018315 | Difference: 142.8180211018315
(INFO) (paste.py) (10-Jan-25 17:14:38) Iteration: 1
(INFO) (paste.py) (10-Jan-25 17:14:38) Solving Pairwise Slice Alignment Problem.
(INFO) (paste.py) (10-Jan-25 17:14:38) Slice 0
(INFO) (paste.py) (10-Jan-25 17:14:39) Slice 1
(INFO) (paste.py) (10-Jan-25 17:14:39) Slice 2
(INFO) (paste.py) (10-Jan-25 17:14:40) center_ot done
(INFO) (paste.py) (10-Jan-25 17:14:40) Solving Center Mapping NMF Problem.
(INFO) (paste.py) (10-Jan-25 17:14:51) Objective 0.895093733781344 | Difference: 143.71311483561286
(INFO) (paste.py) (10-Jan-25 17:14:51) Iteration: 2
(INFO) (paste.py) (10-Jan-25 17:14:51) Solving Pairwise Slice Alignment Problem.
(INFO) (paste.py) (10-Jan-25 17:14:51) Slice 0
(INFO) (paste.py) (10-Jan-25 17:14:51) Slice 1
(INFO) (paste.py) (10-Jan-25 17:14:52) Slice 2
(INFO) (paste.py) (10-Jan-25 17:14:52) center_ot done
(INFO) (paste.py) (10-Jan-25 17:14:52) Solving Center Mapping NMF Problem.
(INFO) (paste.py) (10-Jan-25 17:15:03) Objective 0.9025986308668262 | Difference: 0.007504897085482187
(INFO) (paste.py) (10-Jan-25 17:15:03) Iteration: 3
(INFO) (paste.py) (10-Jan-25 17:15:03) Solving Pairwise Slice Alignment Problem.
(INFO) (paste.py) (10-Jan-25 17:15:03) Slice 0
(INFO) (paste.py) (10-Jan-25 17:15:03) Slice 1
(INFO) (paste.py) (10-Jan-25 17:15:04) Slice 2
(INFO) (paste.py) (10-Jan-25 17:15:04) center_ot done
(INFO) (paste.py) (10-Jan-25 17:15:04) Solving Center Mapping NMF Problem.
(INFO) (paste.py) (10-Jan-25 17:15:14) Objective 0.8967856411071421 | Difference: 0.0058129897596841396
(INFO) (paste.py) (10-Jan-25 17:15:14) Iteration: 4
(INFO) (paste.py) (10-Jan-25 17:15:14) Solving Pairwise Slice Alignment Problem.
(INFO) (paste.py) (10-Jan-25 17:15:14) Slice 0
(INFO) (paste.py) (10-Jan-25 17:15:15) Slice 1
(INFO) (paste.py) (10-Jan-25 17:15:15) Slice 2
(INFO) (paste.py) (10-Jan-25 17:15:16) center_ot done
(INFO) (paste.py) (10-Jan-25 17:15:16) Solving Center Mapping NMF Problem.
(INFO) (paste.py) (10-Jan-25 17:15:26) Objective 0.9024441130949585 | Difference: 0.005658471987816438
(INFO) (paste.py) (10-Jan-25 17:15:26) Iteration: 5
(INFO) (paste.py) (10-Jan-25 17:15:26) Solving Pairwise Slice Alignment Problem.
(INFO) (paste.py) (10-Jan-25 17:15:26) Slice 0
(INFO) (paste.py) (10-Jan-25 17:15:27) Slice 1
(INFO) (paste.py) (10-Jan-25 17:15:27) Slice 2
(INFO) (paste.py) (10-Jan-25 17:15:27) center_ot done
(INFO) (paste.py) (10-Jan-25 17:15:27) Solving Center Mapping NMF Problem.
(INFO) (paste.py) (10-Jan-25 17:15:37) Objective 0.9052546190584809 | Difference: 0.0028105059635223917
(INFO) (paste.py) (10-Jan-25 17:15:37) Iteration: 6
(INFO) (paste.py) (10-Jan-25 17:15:37) Solving Pairwise Slice Alignment Problem.
(INFO) (paste.py) (10-Jan-25 17:15:37) Slice 0
(INFO) (paste.py) (10-Jan-25 17:15:38) Slice 1
(INFO) (paste.py) (10-Jan-25 17:15:38) Slice 2
(INFO) (paste.py) (10-Jan-25 17:15:38) center_ot done
(INFO) (paste.py) (10-Jan-25 17:15:38) Solving Center Mapping NMF Problem.
(INFO) (paste.py) (10-Jan-25 17:15:50) Objective 0.9018425708988059 | Difference: 0.0034120481596749963
(INFO) (paste.py) (10-Jan-25 17:15:50) Iteration: 7
(INFO) (paste.py) (10-Jan-25 17:15:50) Solving Pairwise Slice Alignment Problem.
(INFO) (paste.py) (10-Jan-25 17:15:50) Slice 0
(INFO) (paste.py) (10-Jan-25 17:15:50) Slice 1
(INFO) (paste.py) (10-Jan-25 17:15:50) Slice 2
(INFO) (paste.py) (10-Jan-25 17:15:51) center_ot done
(INFO) (paste.py) (10-Jan-25 17:15:51) Solving Center Mapping NMF Problem.
(INFO) (paste.py) (10-Jan-25 17:16:03) Objective 0.9057043130804422 | Difference: 0.003861742181636263
(INFO) (paste.py) (10-Jan-25 17:16:03) Iteration: 8
(INFO) (paste.py) (10-Jan-25 17:16:03) Solving Pairwise Slice Alignment Problem.
(INFO) (paste.py) (10-Jan-25 17:16:03) Slice 0
(INFO) (paste.py) (10-Jan-25 17:16:04) Slice 1
(INFO) (paste.py) (10-Jan-25 17:16:04) Slice 2
(INFO) (paste.py) (10-Jan-25 17:16:04) center_ot done
(INFO) (paste.py) (10-Jan-25 17:16:04) Solving Center Mapping NMF Problem.
(INFO) (paste.py) (10-Jan-25 17:16:18) Objective 0.9015663963138815 | Difference: 0.004137916766560723
(INFO) (paste.py) (10-Jan-25 17:16:18) Iteration: 9
(INFO) (paste.py) (10-Jan-25 17:16:19) Solving Pairwise Slice Alignment Problem.
(INFO) (paste.py) (10-Jan-25 17:16:19) Slice 0
(INFO) (paste.py) (10-Jan-25 17:16:19) Slice 1
(INFO) (paste.py) (10-Jan-25 17:16:19) Slice 2
(INFO) (paste.py) (10-Jan-25 17:16:20) center_ot done
(INFO) (paste.py) (10-Jan-25 17:16:20) Solving Center Mapping NMF Problem.
(INFO) (paste.py) (10-Jan-25 17:16:31) Objective 0.9040103819191492 | Difference: 0.0024439856052677067
(INFO) (paste.py) (10-Jan-25 17:16:31) Center slice computed.
(INFO) (dataset.py) (10-Jan-25 17:16:31) Center aligning
(INFO) (dataset.py) (10-Jan-25 17:16:31) Stacking slices around center slice
We can render the center slice and the aligned dataset as usual.
[8]:
center_slice
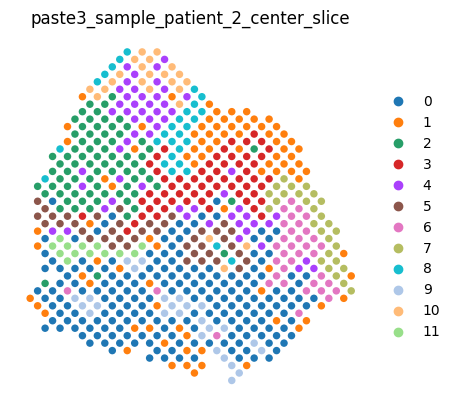
[8]:
<paste3.dataset.Slice at 0x7fabab7ae570>
[9]:
aligned_dataset
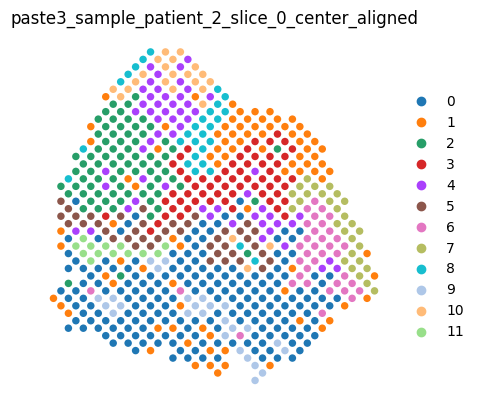
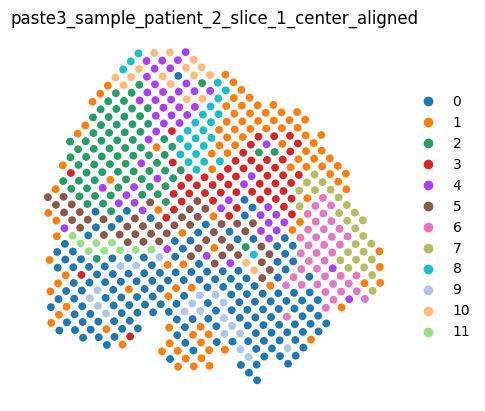
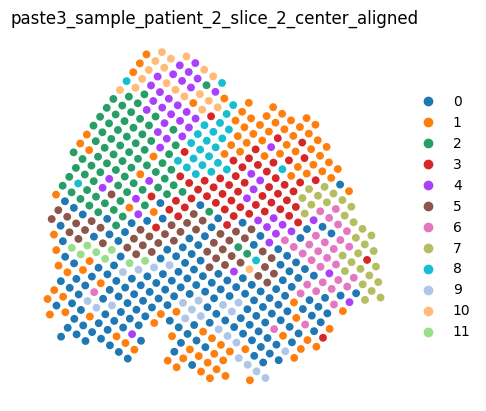
[9]:
<paste3.dataset.AlignmentDataset at 0x7fabb9c8c9b0>
Pairwise aligning a Dataset
A dataset can be pairwise aligned using the <dataset>.pairwise_align method. An overlap_fraction value (between 0 and 1) can be specified.
A value of None results in pairwise alignment that is identical to the approach mentioned in the Paste paper. Any other value between 0 and 1 results in pairwise alignment explained in the Paste2 paper.
[10]:
pairwise_aligned_dataset = dataset.pairwise_align(overlap_fraction=0.7)
(INFO) (dataset.py) (10-Jan-25 17:16:33) Finding Pi for slices 0 and 1
(INFO) (paste.py) (10-Jan-25 17:16:33) GPU is not available, resorting to torch CPU.
(INFO) (dataset.py) (10-Jan-25 17:16:34) Finding Pi for slices 1 and 2
(INFO) (paste.py) (10-Jan-25 17:16:34) GPU is not available, resorting to torch CPU.
[11]:
pairwise_aligned_dataset[0]
[11]:
<paste3.dataset.AlignmentDataset at 0x7fabbc356870>