
Converts mutation data stored as dictionaries into a bipartite NetworkX graph.
| Parameters: |
|
|---|
For more information on the internal format of mutation data used by Multi-Dendrix, see multi_dendrix.load_mutation_data().
| Returns: | Bipartite NetworkX graph G=(V, E) with genes and patients as nodes, and edges representing a mutation in a particular gene in a particular patient. |
|---|
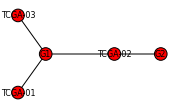
>>> import networkx as nx
>>> mutation2patients = {"G1" : ["TCGA-01", "TCGA-02", "TCGA-03"], "G2" : ["TCGA-02"]}
>>> patient2mutations = {"TCGA-01" : ["G1"], "TCGA-02" : ["G1", "G2"], "TCGA-03" : ["G1"]}
>>> G = construct_mutation_graph(mutation2patients, patient2mutations)
>>> nx.draw_spectral(G)
See also: graph_to_mutation_data(), permute_mutation_data().