
Permutes the given mutation data stored in bipartite graph G=(V, E) by performing | E | * Q edge swaps.
| Parameters: |
|
|---|---|
| Returns: | Permuted version of G reformatted into the mutation data format used by Multi-Dendrix (see graph_to_mutation_data() and multi_dendrix.load_mutation_data()). |
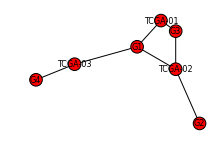
>>> import networkx as nx
>>> G = nx.Graph()
>>> G.add_edges_from([("G1", "TCGA-01"), ("G1", "TCGA-02"), ("G1", "TCGA-03"),
("G2", "TCGA-02"), ("G3", "TCGA-01"), ("G3", "TCGA-02"), ("G4", "TCGA-03")])
>>> nx.draw_spectral(G, dpi=72, node_size=125, font_size=8)
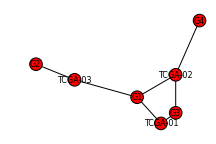
>>> M = permute_mutation_data(G, ["G1", "G2", "G3", "G4"], ["TCGA-01", "TCGA-02", "TCGA-03"])
>>> M
(4, 3, ['G4', 'G3', 'G2', 'G1'], ['TCGA-03', 'TCGA-02', 'TCGA-01'],
{'G4': set(['TCGA-02']), 'G3': set(['TCGA-02', 'TCGA-01']), 'G2': set(['TCGA-03']),
'G1': set(['TCGA-03', 'TCGA-02', 'TCGA-01'])},
{'TCGA-03': set(['G2', 'G1']), 'TCGA-02': set(['G4', 'G3', 'G1']),
'TCGA-01': set(['G3', 'G1'])})
>>> H = construct_mutation_graph(M[-1], M[-2])
>>> nx.draw_spectral(H, dpi=72, node_size=125, font_size=8)
See also: construct_mutation_graph(), graph_to_mutation_data().